Research Topics in the Field of Bioinformatics
March 27, 2023 2023-04-13 1:33Research Topics in the Field of Bioinformatics
From Genome to Drug
Bioinformatics played a decisive role in the most prominent scientific achievements during recent years, the sequencing of the human genome. With the sequence being known, the annotation of the genome can begin.

This means searching for genes in DNA , identifying the corresponding gene products (proteins, RNA), as well as determining their structure and function.
To fully understand protein function, one has to consider their interplay. These interactions are represented by metabolic and regulatory networks which, among other things, allow for simulating metabolic processes. Bioinformatics methods are used to find the reasons for genetically caused diseases to find new possibilities for therapies.
In general, drugs act by influencing the proteins that are involved in metabolism. Based on the assembly of the human genome, bioinformatics methods allow researchers to find proteins (Targets) that are better suited for treating certain diseases.
Bioinformatics delivers important contributions to the development of new drugs. Databases enable the search through large amounts of data in order to find new candidate drugs (screening, docking), that are efficient, have fewer side-effects and are capable of reaching the right destination in the body (crossing of biological barriers).
Bioinformatics supports the optimization of known therapies. The comparison of complete genomes of different individuals makes it possible to trace differences (i.e. SNPs), which may play a role when deciding on the individual therapy for a patient.
Viral infections present a great challenge for drug development and therapy. The fact that viruses like HIV show high genomic variability, can result in the occurrence of viral mutations that confer resistance to the prescribed drugs.
Therefore a physician is faced rather frequently with the problem of finding a new therapy for each patient infected with a particular strain. Bioinformatics methods have been developed to understand the relationship between viral mutations and drug resistance, leading to better therapeutic strategies.
Genome Sequencing
The human genome consists of 46 long DNA molecules (chromosomes) contained within the nucleus of the cell. The chromosomes carry genetic information.
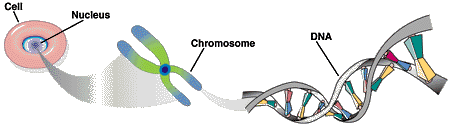
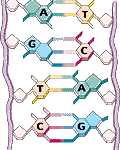
Each DNA molecule consists of two strand in the form of a double helix. Each DNA strand is a linear polymer that consists of similar subunits (monomers) connected end to end.
Within each monomer one can find a sugar, a phosphate and a base component. The sequences of bases represents a form of linear infomation. There are four bases denoted by the letters A, C, G and T. The bases A,T and G,C are complementary, i.e. bind to each other. Based on this base pair complementarity a single strand contains the full genetic information.
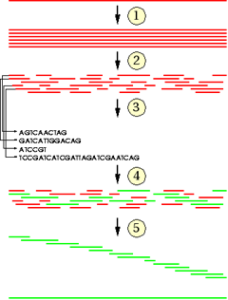
The goal of sequencing is to obtain the ordered set of bases contained in the DNA in form of a long string. The sequencing machines cannot read the whole genome in one step. Therefore, the genome has to be cut into smaller pieces. In order to be able to reassemble the pieces they have to be overlapping.
This can be achieved by generating many copies of a DNA strand and cutting it into pieces randomly (with high pressure, ultrasound).
In the process of sequence assembley the full sequence of nucleotides is gathered from overlap information by performing a stepwise search for pieces with overlapping ends.
Then overlapping pieces are put together.
Bioinformaics provides suitable algorithms for the assembley step. These algorithms have to be very efficient as the number of pieces and hence the number of pairwise comparisons for overlaps is large. In addition the algorithms have to deal with such problems as repetitive sequences or reading errors in the genome pieces.
Gene Annotation
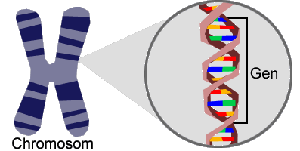
The chromosomes contain the genome of every organism. The genome contains sequence regions (genes) that code for proteins and other molecular constituents.
The proportion of coding sequence in relation to the total genome is rather small.
After sequencing a genome the task is to localize the genes. This step, part of the annotation process, requires bioinformatics methods such as pattern recognition and sequence alignment.
Structure and Function of Gene Products
Once a gene has been found, the aim is to elucidate the structure and function of the building block encoded by the gene. The sequence of a gene determines the spatial structure of the building block.
This three-dimensional shape, in turn, influences which task the building block in question performs in the body. Bioinformatics develops both algorithms that allow the spatial structure of a building block to be predicted and methods for functional analysis.

Since the sequence determines the structure of a building block, it makes sense to search for similar sequences in databases in order to transfer knowledge about the function of already known building blocks to newly sequenced genes (sequence alignment). The function of building blocks can also be determined by comparing their spatial structure.
One promising approach to elucidating function is the use of so-called DNA chips or the measurement data generated with the help of this technique: DNA chips make it possible to obtain information about which proteins are active in the cell at a given time and which are not. In this way, one can determine information about the probable function of proteins.

Metabolic Pathways
After elucidating the function of individual proteins, it is of particular interest to study the metabolic pathways of an organism.
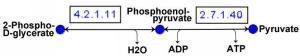
A metabolic pathway is a sequence of reactions that together provide for the conversion of one substance into another. The individual reactions are essentially characterized by the catalyzing enzyme (the enzyme that accelerates the reaction).
Important examples of metabolic pathways are the citrate cycle or glycolysis. The totality of all pathways forms a metabolic network.
In addition to metabolic pathways, there are equally important regulatory pathways that organize communication between cells and the overall control of processes in the organism. With the help of a wide variety of signals and communication mechanisms, the processes in the organism can be specifically controlled. All regulatory pathways together form the regulatory network of an organism.
In order to be able to analyze both metabolic and regulatory networks with the help of the computer, appropriate data models are required. The goal is to find a suitable model that can represent both types of networks.
The network representations form the basis for developing new tools that are needed, for example, for target identification, drug design or elucidating the causes of genetic diseases.
In basic research, these networks can be used to compare metabolic processes of different organisms. For example, information about the metabolism of one organism can be used to understand the newly sequenced genome (and thus the metabolic pathways) of another organism.
Target Identification
With the help of metabolic and regulatory networks, scientists can start to understand the metabolism in the human organism. They are particularly interested in disturbances of the normal processes, since these are often the cause of diseases.
If one knows where the cause of a disease lies, for example, one can use the metabolic networks to carry out a more detailed analysis and look for “points of attack” (hence “target” identification) for a possible treatment. The targets sought are usually enzymes that control certain reactions in the metabolism.
Once an enzyme has been identified as the “culprit,” an attempt can be made to influence the activity of this enzyme with certain active substances in order to remedy the cause of the disease. The use of the previously modeled networks also makes it possible to analyze possible side effects on other processes in the body that should not be influenced by treatment.
Drug Design
The identification of a target alone is not enough for effective treatment – a suitable active ingredient must first be developed. Such an active ingredient must influence the disease-causing enzyme in such a way that it no longer interferes with the metabolism.
This can be done by inhibiting the activity of the enzyme. If a disease arises because there is a deficiency of a certain substance, an active ingredient can also be designed in such a way that it stimulates the production of an enzyme, which in turn then controls the necessary (and previously absent or insufficient) metabolic processes.
At the molecular level, influencing enzyme activity by an active substance means “docking” the substance to the target protein (target) in order to exert its effect in this way.
For this reason, bioinformatic methods have been developed that virtually screen the target for ligands that can bind to and inhibit the protein. Another option is to find other proteins that regulate the activity of the target through binding and complex formation. In both cases, docking is the main technique.
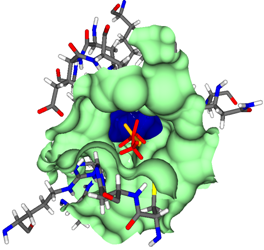
Docking
Proteins can be roughly thought of as a long chain of amino acids. Each amino acid consists of a functional group and the so-called residue. The residues of the amino acids are also called side chains in the protein. The chain formation occurs at the functional group where the individual proteins form a peptide bond. This chain formation can theoretically be continued for any length of time. The different amino acids are generally abbreviated with a 3-letter code. If one wants to describe a protein, it is sufficient to write down this sequence of amino acids.
The theoretical determination of the interactions between two proteins or protein and ligand, roughly speaking the binding, is of great interest because it allows the verification of hypotheses during the so-called drug design without having to rely on real laboratory experiments.
A recurring problem is the so-called protein-protein docking, which is defined as follows:
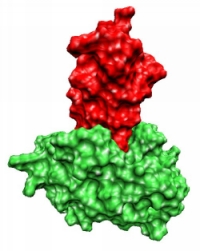
Let the spatial structures of proteins A and B be given, which are known to form a complex AB, then protein-protein docking is understood to correctly predict the complex structure AB.
There are different approaches to protein-protein docking. The simplest approach considers proteins as rigid bodies. This is called rigid body docking (RBD). This approach goes back to the well-known key-lock principle proposed by Emil Fischer in 1894.
This model states that the proteins to be docked, in his case enzyme and matching substrate, are rigid bodies that must have geometrically complementary regions. Although proteins are not rigid bodies and have mobile side chains, studies showed that the backbone, i.e., the basic structure consisting of the functional groups linked by peptide bonds, hardly changes in many proteins during complex formation.
As a rule, docking yields not just one drug candidate, but a large number that meet the purely physical requirements to interact with the target. From the set of these candidates, those that can be considered as an effective drug must now be selected. Potential problems can arise, for example, if the developed active ingredient does not find its way to its target in the first place, as it is exposed to a variety of different environments and biological barriers in the human body.
Biological Barriers
As soon as a potential active ingredient is found, it must be brought to the site of action. To do this, a number of biological barriers always have to be overcome. The first barriers already appear when the substance is absorbed into the body. Here, the intestinal wall or the membrane of the alveoli must be penetrated before the active substance is decomposed or excreted.
In addition, transport losses must be taken into account. Even if one only wants to interfere with very primitive (unicellular) organisms, at least cell membranes have to be overcome. At this stage, practical experimentation is particularly necessary – a complex and expensive procedure for testing the suitability of an active substance.
Much research is still needed in this subfield, especially for bioinformatics, to provide alternative methods that can help save time and money.

