Bioinformatics (Prof. Helms)
October 21, 2022 2023-04-13 1:33Bioinformatics (Prof. Helms)
About us
The research group headed by Prof. Dr. Helms applies numerical simulations, as well as statistical methods to study biomolecular interactions and control loops in cells. Current topics of interest are the modeling of protein-protein contacts, as well as interactions between DNA and proteins, which both play a central role in the control of all major biological processes. Within the field of membrane bioinformatics, we are deriving structural and functional motifs from the protein sequences of integral membrane proteins. Of particular interest to us is to increase the understanding of specific molecular interactions in the context of large scale biological processes. This affords insights into the origin of diseases at the molecular level.
Head of the Group

Prof. Dr. Volkhard Helms
Volkhard Helms has been Professor of Bioinformatics in the Center of Bioinformatics Saar since 2003 and is also a member of the CBI Saar Directorate. He studied physics in Freiburg and Munich and obtained his PhD at the European Molecular Biology Laboratory in Heidelberg in 1996. He then carried out post- doctoral work at the University of California, San Diego, and went on to lead a research group in theoretical biophysics at the Max Planck Institute of Biophysics in Frankfurt. In 2001 he was selected as an EMBO Young Investigator.
"Via the modelling of single biomolecular interactions and regulation processes the group desires to arrive at a comprehensive understanding of cell biological processes."
Our Projects
Functional assignment of transmembrane proteins
About a quarter of all proteins is integrated into the cell membrane or into the membranes of cellular compartments. There, they mediate the transport of small molecules through the membrane, or transduct signals from messenger substances, which dock to the cell into its interior. We are particularly interested in elucidating the assignment of substrate molecules to their corresponding membrane transporters, because only little is known so far. For this purpose, we are developing new statistical methods, as well as a software environment to model a virtual cell membrane.
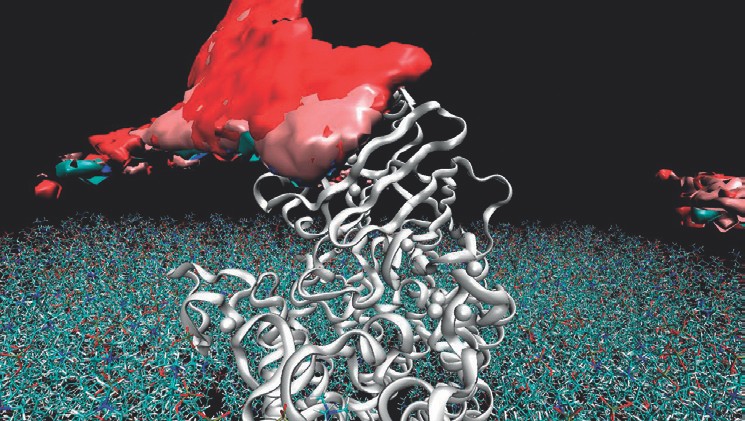
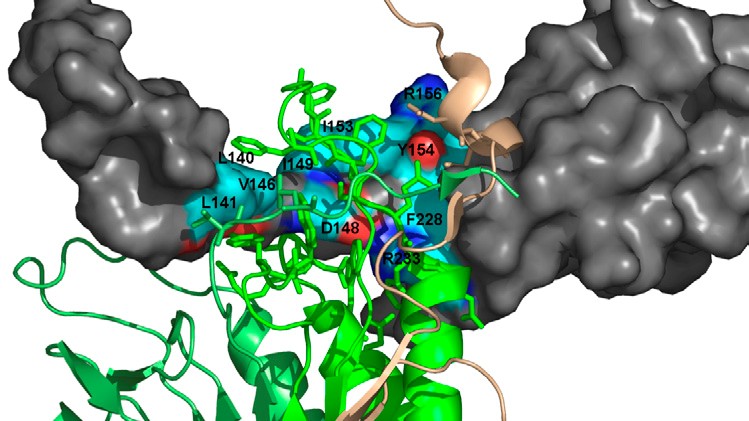
Mediation of protein-protein interactions
Within the cell the majority of proteins is in permanent exchange with other proteins. We use complex computer simulations to elucidate these associations in atomistic details. We found out that the surrounding water has a pronouncing effect during aggregation. Based on these observations, we now investigate the role of small ligand molecules on such protein-protein interactions, which can either be inhibited or in turn facilitated. This strategy allows to develop new clinical therapies for the treatment of cancer.